Input
You have 2 choices of starting point.
- Either you input a sequence and let our tool provide you with a multiple sequence alignment (MSA), or
- you already have the MSA and input that.
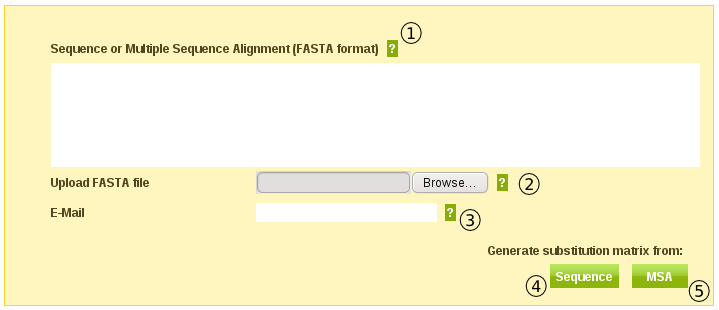
A sequence or MSA can be pasted in the text area (1) in the input form (Fig. 1) or you can use the file upload (2).
Some of the calculations will take some time to complete. By entering your email address in the email field (3) you will receive an email with a URL to your results.
If you want the tool to generate a conserved block MSA from your input click the "Sequence" button (4).
If you have given a conserved block MSA as input you can click the "MSA" button (5).
Fig. 1
Generate Multiple Sequence Alignment
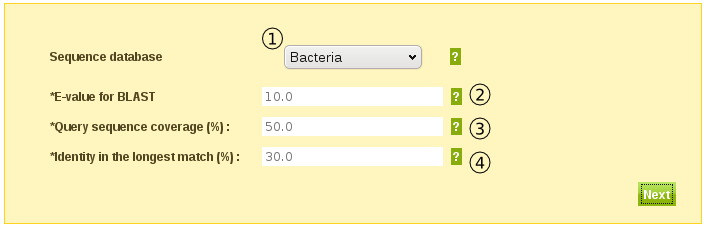
If you chose to enter a sequence in the step above, the tool will in this step produce a multiple sequence alignment for you based on the hits it get against the database you chose (1 in fig.2).
You can influence the number of hits by changing the following parameters
- E-value (2) - Minimum significance value of search hit against your query sequence.
- Coverage (3) - Minimum percentage overlap between your query sequence and a search hit sequence.
- Identity (4) - Minimum percentage of sequence identity between your query sequence and a search hit, in the longest match block of the pairwise alignment of the two.
Fig. 2
Generate Substitution Matrix
In this step the tool will generate the substitution matrix and display the results.
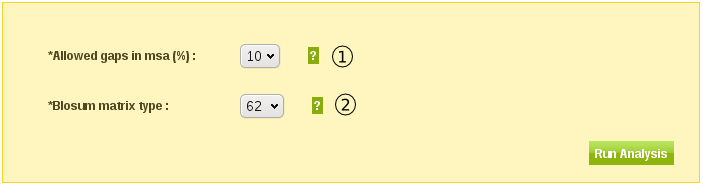
The "Allowed gaps in MSA" (1 in fig. 3) parameter tells the tool to calculate substitution rates for only those positions (columns) in the MSA that have maximum x percent of gaps.
By default the tool allows 10% of gaps in the positions to calculate substitution rates for.
The "Blosum matrix type" parameter specifies which evolutionary distance to use in the preprocessing of the actual substitution rate calculations.
E.g. for BLOSUM62, sequences in the MSA with more than 62% sequence identity are clustered together as one entity before the final substitution rate calculation starts.
The default is BLOSUM62.
Fig. 3
Example Output
Click Here to see a sample Example Output