Background
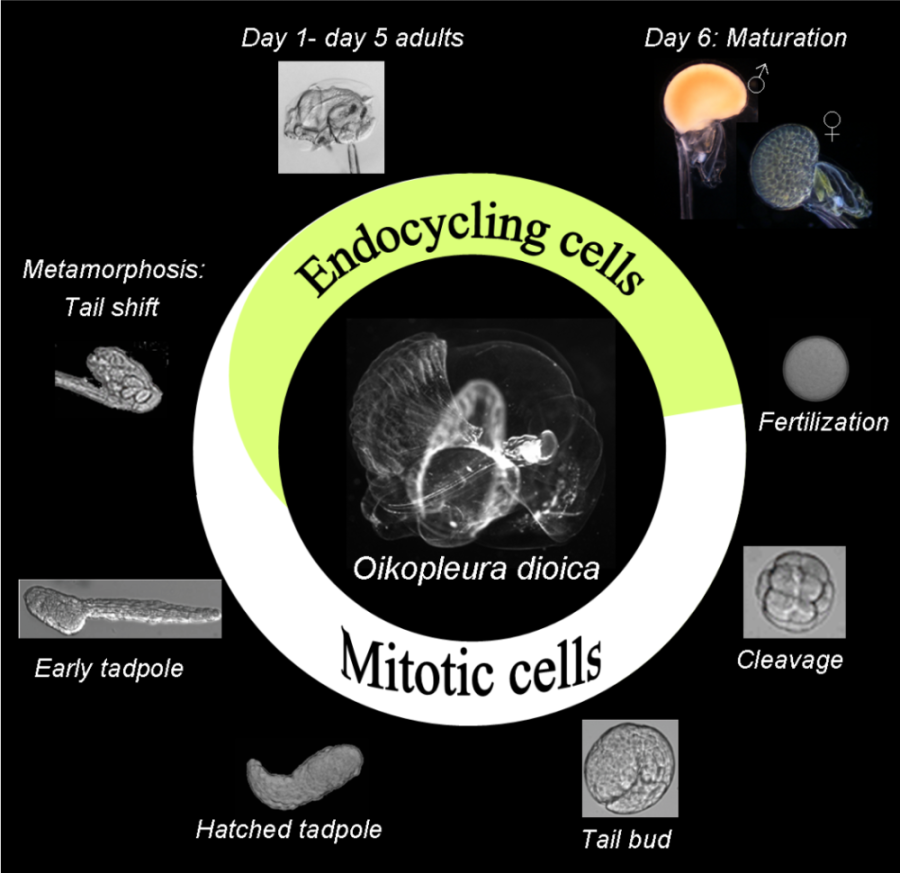
Life cycle figure: The appendicularian Oikopleura dioica is an abundant pan-global zooplankton that retains a tadpole form throughout its life cycle.
The urochordates are the closest living relatives of vertebrates. Oikopleura dioica, a representative of the larvacean class has an extremely short life cycle of 6 days in culture and the smallest genome identified in chordates (reviewed in Nishida 2008). Oikopleura shows interesting life history features concerning histone gene organization and regulation, as well as the spatial and temporal use of histone variants (Moosmann et al., in preparation). Histone variants alter the composition of individual nucleosomes and expand the repertoire of histone posttranscriptional modifications. The exchange of conventional histones for histones variants can affect gene expression, centromere function and is supposably involved in establishing epigenetic marks in the male germ line. The Oikopleura histone complement is very compact with relatively few genes encoding at least the same amount of histone isoforms reported for mammals and other organisms. The manageable number of histone genes and the different cell cycle types, prominent in the Oikopleura life cycle, make it a very suitable model to study histone variant expression and their post-translational modifications in mitotic, endocycling and meiotic nuclei.
The life cycle of this marine urochordate is characterized by a developmental switch between two different cell cycle types. In early embryonic development, a fixed number of cells is established through rapid mitotic divisions. At metamorphosis (tail shift), cells in most tissues exit mitosis and increase their nuclear volume by endoreduplication. In the epithelium of adult animals that secretes the mucous house, this results in different field specific ploidy levels (Ganot and Thompson, 2002). As the sole dioecious species reported in tunicates, ovaries of mature females show similarities to the meroistic syncitium of Drosophila, where proliferating germline nuclei occupy a common cytoplasm and give rise to two types of nuclei: meiotic nuclei and endocycling nurse nuclei (Ganot et al, 2007a,b, 2008).
The Oikopleura Histone Webpage summarizes all histone sequence- and expression data for this urochordate. Histone genes in Oikopleura are clustered in small groups dispersed throughout the genome as shown in the gene maps. In total we have identified 47 histone genes (10 H3, 6 H4, 15 H2A , 11 H2B and 5 H1 genes) in the latest draft genome sequence (Genoscope) of Oikopleura encoding 32 different histone proteins ( 7 H3, 5 H1, 2 H4, 7 H2B and 11 H2A). Expression profiles for the histone genes throughout Oikopleura development were assessed by quantitative PCR. With few exceptions, Oikopleura histone genes are co-regulated in clusters and several variants are exclusively expressed at distinct developmental stages. Histone expression during development can be assigned to five different expression profiles: Expression during the entire life cycle ("A": all stages), Expression at organogenesis ("H": hatched embryos), expression peaking at metamorphosis ("T": tail shift), expression peaking in juveniles at day 3 and day 4 (D3/D4) and male specific expression (M). For further information about the Oikopleura histone complement and generation of data see also (Moosmann et al., in preparation). User Information: Please note that the scaffold names referring to the Genoscope draft genome sequences may not be definitive. The sex specific expression data presented on this webpage was generated from whole animal cDNA and not from female/male gonad extracts (for a more detailed description please see Moosmann et al., in preparation). For a few genes that lack specific primer pairs the expression data is not presented.
User Information
Please note that the scaffold names referring to the Genoscope draft genome sequences may not be definitive. The sex specific expression data presented on this webpage was generated from whole animal cDNA and not from female/male gonad extracts (for a more detailed description please see Moosmann et al., in preparation). For a few genes that lack specific primer pairs the expression data is not presented.
References
- Ganot P, Bouquet JM, Kallesøe T and Thompson EM (2007). The Oikopleura coenocyst, a unique chordate germ cell permitting rapid, extensive modulation of oocyte production. Dev Biol. 302, 591-600. link
- Ganot P, Kallesøe T and Thompson EM (2007). The cytoskeleton organizes germ nuclei with divergent fates and asynchronous cycles in a common cytoplasm during oogenesis in the chordate Oikopleura. Dev Biol.302, 577-590. link
- Ganot P, Moosmann-Schulmeister A and Thompson EM (2008). Oocyte selection is concurrent with meiosis resumption in the coenocystic oogenesis of Oikopleura. Dev Biol. 324, 266-276. link
- Nishida H (2008). Development of the appendicularian Oikopleura dioica: culture, genome, and cell lineages.Dev Growth Differ. 50, 239-256. link
Contact
If you have any questions about the Oikopleura Histone Database please send an email to Alexandra Moosmann or Eric Thompson
For more information about the Thompson Group go here